SimPATHy
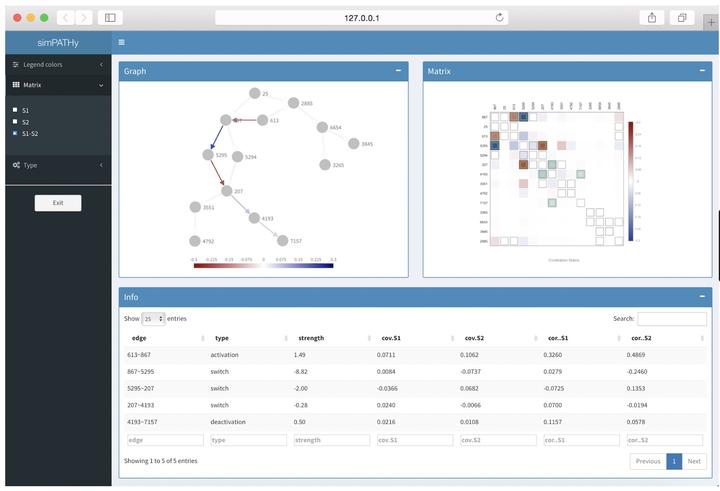
simPATHy is a model for simulating data from perturbed biological pathways. It is implemented as an R package and produces synthetic datasets useful to validate GSA methods focused on topological properties of biological networks.
Given a graph structure representing a biological network, functions in simPATHy allow simulating data in two different conditions. The two conditions, assumed to share the same graphical structure, are referred to as reference and dysregulated condition. Dysregulation is defined as a change in the strength of the links between network nodes. Such changes can be interpreted as activation/deactivation of network connections.
Elisa Salviato, Vera Djordjilović, Monica Chiogna, Chiara Romualdi, simPATHy: a new method for simulating data from perturbed biological PATHways, Bioinformatics, Volume 33, Issue 3, 1 February 2017, Pages 456–457, https://doi.org/10.1093/bioinformatics/btw642