BrewerIX
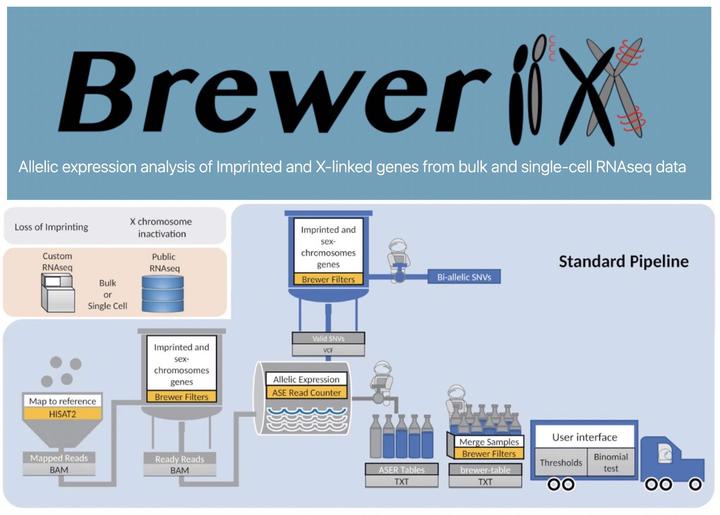
BrewerIX is a standardized approach for the analysis of known imprinted and X-linked genes. Differently from other tools, its implementation strategy allows the user (i) to perform fast and efficient analyses from raw data to the final plots on a standard desktop or laptop computer without requiring any programming skills, (ii) to have comprehensive information of imprinted and X-linked genes taken from different databases that have been manually curated to avoid results misinterpretation, (iii) to graphically visualize the results in an easy and intuitive way. All these features will guarantee the reproducibility of results and transparency.
BrewerIX is implemented as a native graphical application for Linux and macOS. It takes as input either bulk or single-cell RNA-seq data (fastq files), analyzes reads mapped over the SNVs distributed on imprinted genes (see “Knowledge base” section for details), X chromosome and Y chromosome, and generates imprinting and XCI profiles of each sample.
Martini P, Sales G, Diamante L, Perrera V, Colantuono C, Riccardo S, Cacchiarelli D, Romualdi C, Martello G. BrewerIX enables allelic expression analysis of imprinted and X-linked genes from bulk and single-cell transcriptomes. Commun Biol. 2022 Feb 17;5(1):146. doi:10.1038/s42003-022-03087-4.